Answer:
Option D
Neither gene is significantly different
Step-by-step explanation:
Tan Spore gene
Cross-over
Observed=189
Expected=177
Observed-Expected=189-177=12
Standard deviation,
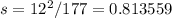
Non-cross-over
Cross-over
Observed=139
Expected=151
Observed-Expected=139-151=-12
Standard deviation,
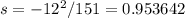
Chi-square value=0.813559+0.953642=1.767202
Rounding off to three decimal places, chi-square value is 1.767
Gray Spore Gene
Cross-over
Observed=204
Expected=216
Observed-Expected=204-216=12
Standard deviation,
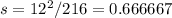
Non-cross-over
Cross-over
Observed=124
Expected=112
Observed-Expected=124-112=12
Standard deviation,
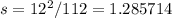
Chi-square value=0.666667+1.285714=1.952381
Rounding off to three decimal places, chi-square value is 1.952
Degree of freedom=n-1 where n is sample size, the sample size here are 2 hence degree of freedom is 2-1=1
From chi-square table, when degree of freedom is 1, chi-square value is 3.841
Since for both genes, the chi-square values are less than 3.841 then we should accept that data since it’s not significantly different